A record is assigned to bin k if k is the bin of the smallest size that fully contains the record. This makes it possible to display variants from files that are so large that the connection to UCSC would time out when attempting to upload the whole file to UCSC. Parameters for VCF custom track definition lines All options are placed in a single line separated by spaces lines are broken only for readability here: In principle, bins can be selected freely as long as each record can be assigned to a bin. Thus in the index file, we do not need to keep the virtual file offset of each record, but only to keep the start offset of a chunk of records assigned to the same bin. Coordinates specified in this region format are 1-based and inclusive. It is a valuable tool for researchers who frequently perform interval queries, for graphical viewer developers who want to display large data files in limited memory, and for genome database developers who intend to support large remote custom tracks. 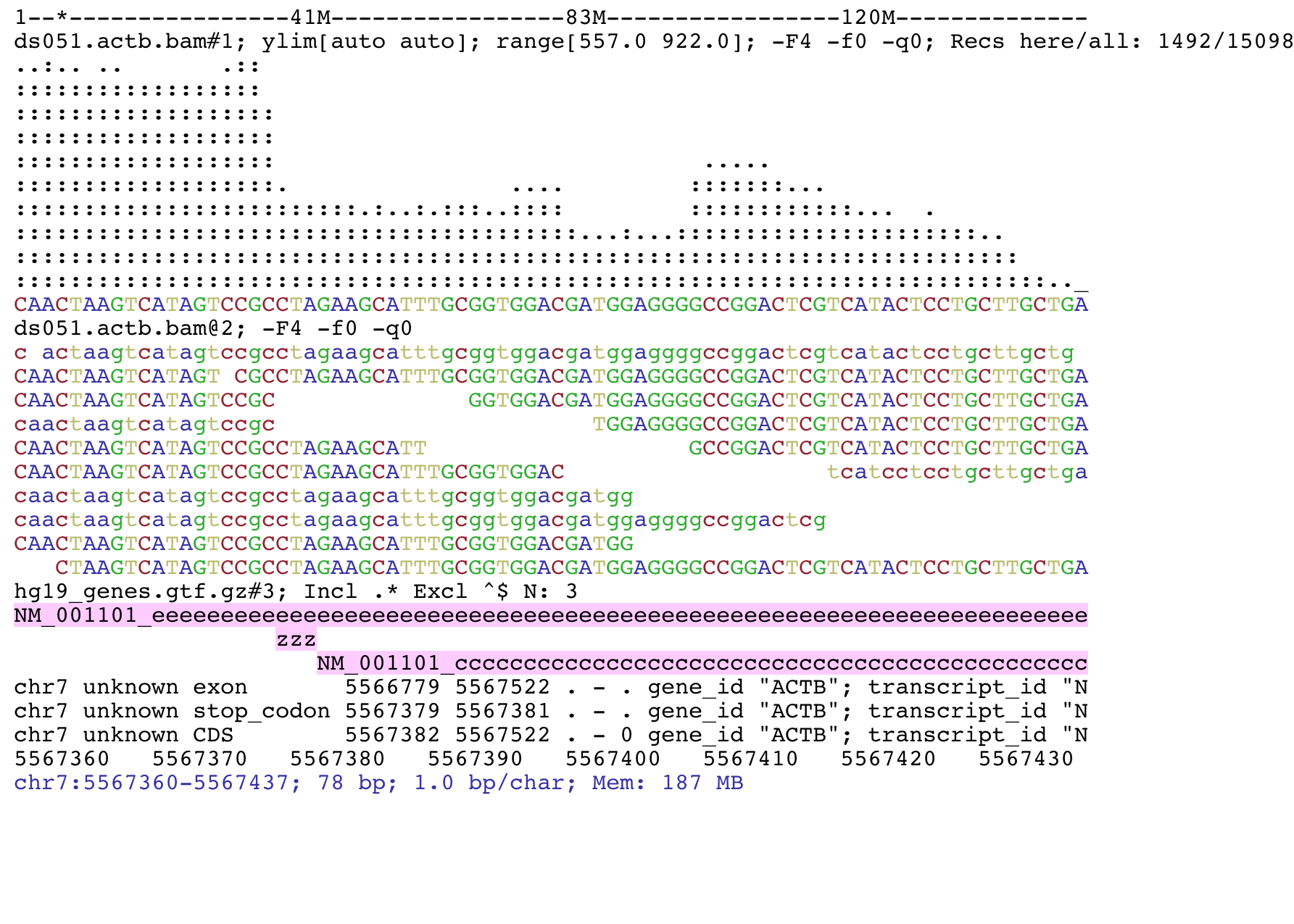
| Uploader: | Mehn |
| Date Added: | 7 December 2015 |
| File Size: | 44.5 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 54483 |
| Price: | Free* [*Free Regsitration Required] |
Variant Call Format VCF is a flexible and extendable line-oriented text format developed by the Genomes Project for releases of single nucleotide variants, indels, copy number variants and structural variants discovered by the project.
If you would like to share your VCF data track with a colleague, learn how to create a URL by looking at Example 6 on the custom tracks page. One can put the data file and the index at an FTP or HTTP server, and other users or even web services will be able to get a slice without downloading the entire file.
It is particularly useful for manually examining local genomic features on the command line and enables genome viewers to support huge data files and remote custom tracks over networks. When a VCF file is compressed and indexed using tabixand made web-accessible, the Genome Browser is able to fetch only the portions of the file necessary to display items in the viewed region. Support Center Support Center.
VCF+tabix Track Format
The line breaks inserted here for readability must be removed before submitting the track line: This makes it possible to display variants from files that are so large that the connection to UCSC would time out when attempting to upload the whole file to UCSC. If you haven't done so already, download and build txbix tabix and bgzip programs. In this scheme, bin 0 spans Mb, 1—8 span 64 Mb, 9—72 8 Mb, 73— 1 Mb, — kb and — span 16 kb intervals.
Nonetheless, Tabix is still fast enough for a genome viewer. This article has been cited by other articles in PMC. The human genome browser at UCSC.
A record is assigned to bin k if k is the bin of the smallest size that fully contains the record. The input data file must be position sorted and compressed by bgzip which has a gzip 1 like interface. Test your installation by running tabix with no command-line arguments; it should print a brief usage message. If you do not have access to a web-accessible server and need hosting space for your VCF files, please see the Hosting section of the Track Hub Help documentation.
The "browser" line above is used to view a small region of chromosome 21 with variants from the. UCSC files rather than 1-based.
The basic version of the track line will look something like this: When we search for samtoolz overlapping a query interval, we will know from the index the leftmost record that possibly overlaps the query interval.
It is a valuable tool for researchers who frequently perform interval queries, for graphical viewer developers who want to display large data files in limited memory, and for genome database developers who intend to support large remote custom tracks.
Remember to remove the line breaks that have been added to the track line for readability or, click here for a text version that you tabi paste without editing:. Accurate whole human genome sequencing using reversible terminator chemistry.
For Permissions, please email: Bgzip compresses the data file in the BGZF format, which is the concatenation of a series of gzip blocks with each block holding at most 2 16 bytes of uncompressed data. Parameters for VCF custom track definition lines All options are placed in a single line separated by spaces lines are broken only for readability here: On the custom track management pageclick the "add custom tracks" button if necessary and make sure that the genome is set to "Human" and the assembly is set to "Feb.
When the data files are sorted by coordinates, records assigned to the same bin tend to be adjacent.
samtools/tabix with https support?
The basic idea of binning is to cluster records into large intervals, called bins. Random access can thus be achieved without the help of additional index structures.
As habix works with concatenated gzip files, it can also seamlessly decompress a BGZF file. Tabix is implemented as a free command-line tool as well as a library in C, Java, Perl and Python. The line breaks inserted here for readability must be removed before submitting the track line:. In principle, bins can be selected freely as long as each record can be assigned to a bin.

Комментариев нет:
Отправить комментарий